The Lysak Lab
About us
We investigate genome evolution across the plant kingdom at different levels of complexity:
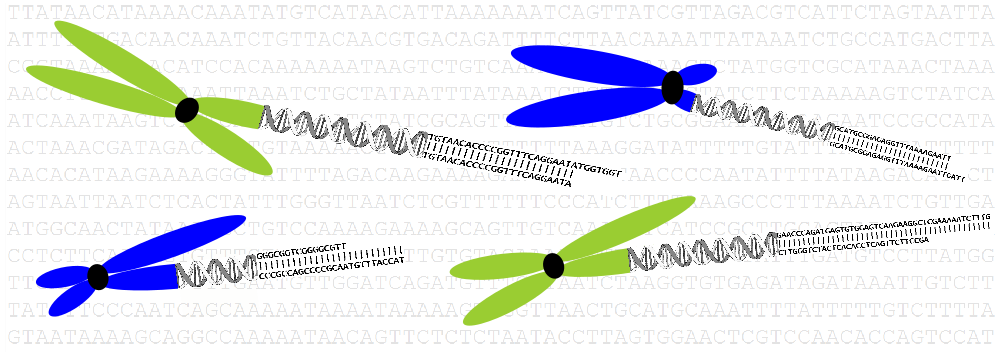
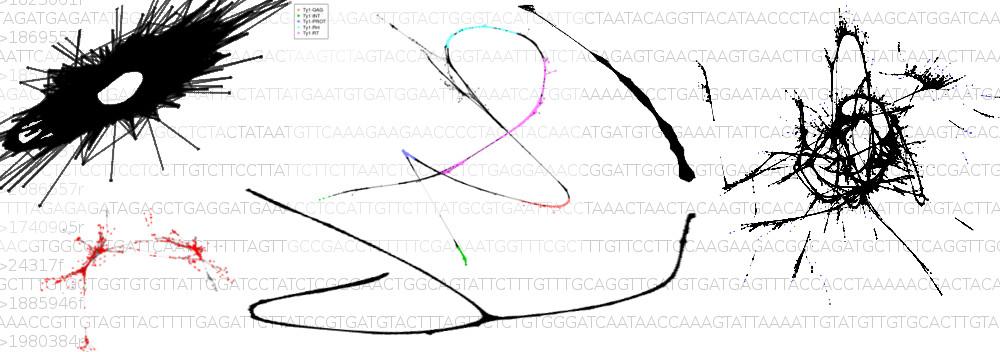
- DNA sequences (NGS-based genome analysis)
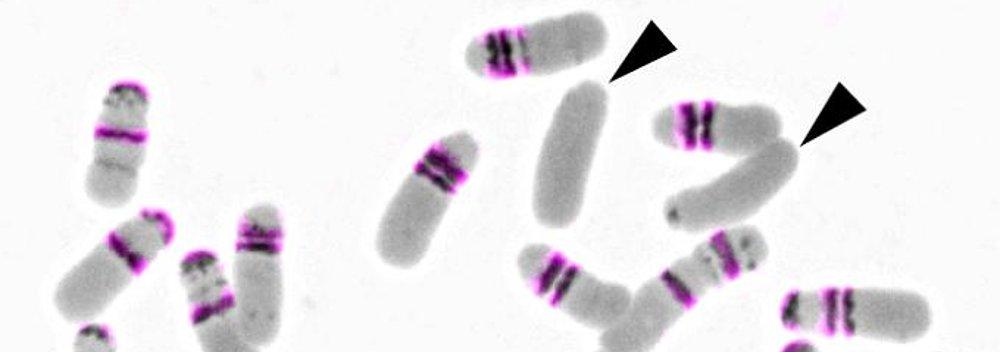
- Chromosomes (chromosome rearrangements, karyotype evolution)
- Genomes (genome collinearity, polyploidy)
- Species (speciation, phylogenomics)
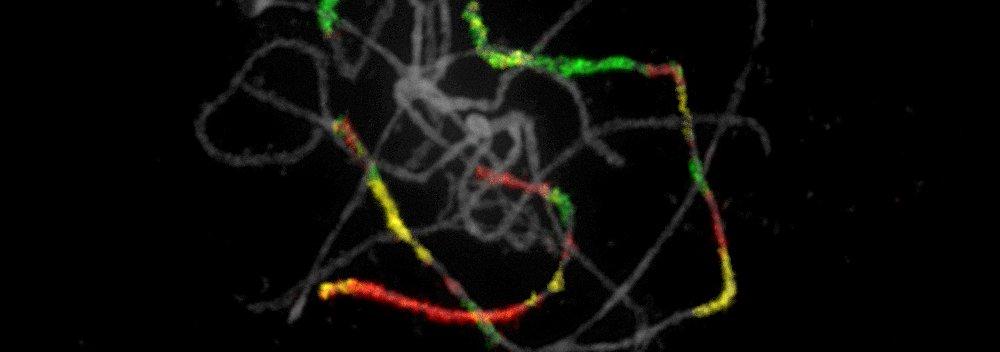
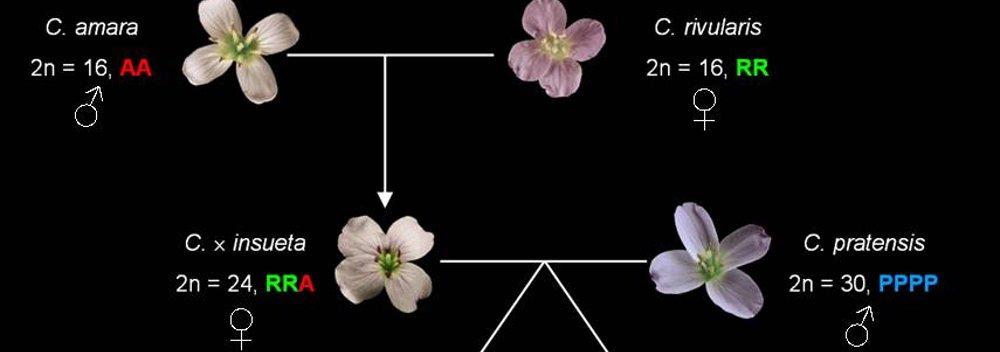
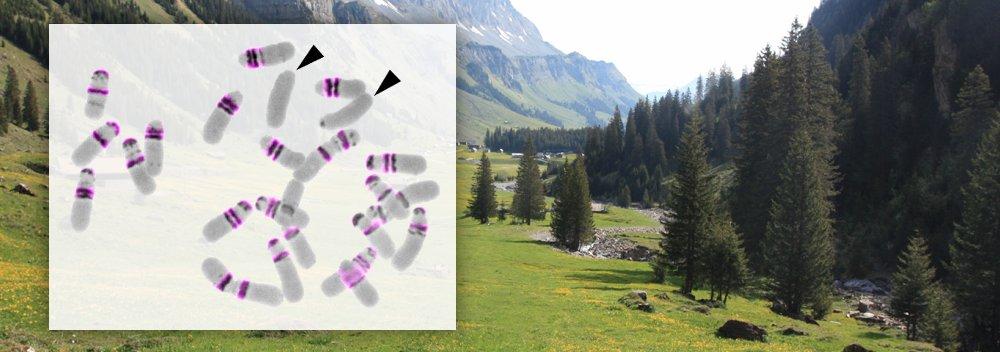
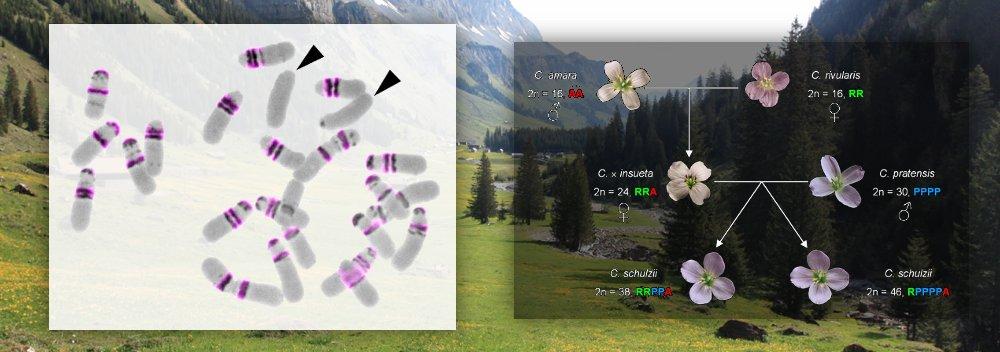
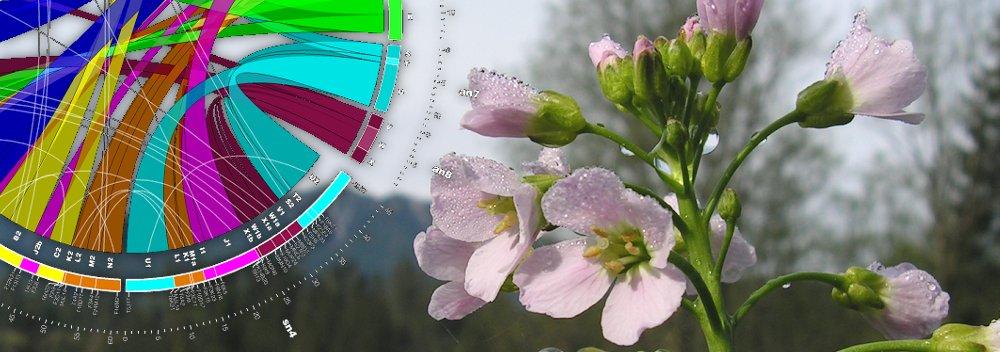
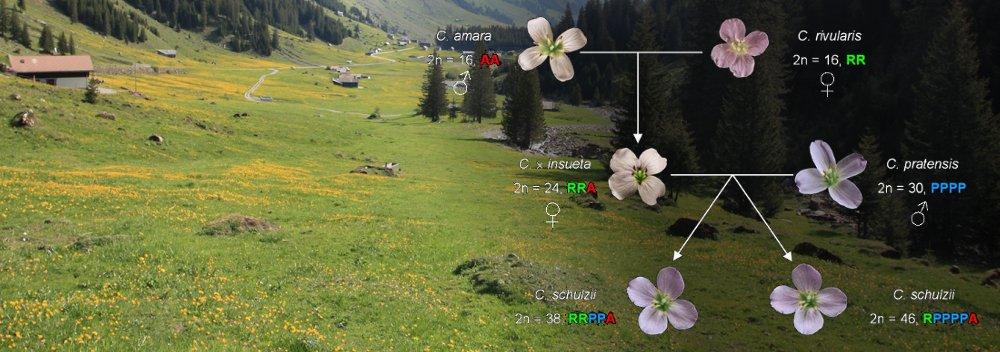
Genome evolution in crucifers (Brassicaceae) is our leading research theme. We are analyzing the incidence, mechanisms and rate of chromosome rearrangements, sequence structure of chromosome breakpoints, modes of centromere shift and elimination, evolution of repetitive sequences, and reconstructing ancestral genomes. We are testing causal links between whole-genome duplication events, karyotypic variation and species radiations.
If you are interested in joining the lab, please contact us!
News
Dr. Yile Huang successfully defended her PhD thesis!
On October 16, Yile successfully defended her PhD thesis entitled Genomics of chromosomal dysploidy in plants. Her primary...
Terezie Malík Mandáková is among this year’s Top Czech Women Scientists selected by Forbes
Congratulations, Teri! More info (in Czech) can be found here: https://forbes.cz...
The largest genome to date in the Brassicaceae decoded!
Martin Lysak, together with the team from Huazhong Agricultural University in China (led by Xianhong Ge), coordinated the sequencing...